Please note that this issue does not affect the analysis itself or the results. Request a quote Existing Customer Login. In many cases, it may suit the analysis requirements to run both tools. Recall that when merging overlapping paired reads, for conflicts the base with the higher quality will be the one found in the merged read, regardless how minor the difference. This issue affects a small subset of variants where all the following apply: Variant calling on results from RNA-Seq Analysis cannot detect insertions and deletions at intron-exon boundaries Issue description Read mappings produced by the RNA-Seq Analysis tool treat deletions and insertions at exon-intron boundaries as splice variants. 
| Uploader: | Vujora |
| Date Added: | 25 August 2005 |
| File Size: | 8.89 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 25580 |
| Price: | Free* [*Free Regsitration Required] |
Please note that this issue does not affect the analysis itself or the results.
Worbench calculation will start. Detailed information about Sequence Lists can be found at the following manual page:. To check if a different Metadata Table was selected, please follow these steps: Unfortunately, it is hard to predict exactly when these hours are, but generally speaking, outside US office hours is probably better than within them.
Filtering on fold change is currently being done for values greater than the value provided in the Wizard.
You may also be interested in the Coverage Analysis tool. The reads are depicted in their forward orientation, which is visualized by the color green. If your reads are as in the 36 nt long example. However, this issue can also affect our internal indexing systems, so we highly recommend that software from affected release lines CLC Genomics Server If you did not choose to keep names on import, then the sequences should be in the same order as in your original fastq file if it was single data.
An ambiguity code will be assigned to any base where the two members of a pair of reads overlap, but genomjcs not agree on the base call. For users of the Genomics Workbench, there are three phases when running a de novo assembly: Some MNVs fitting genlmics description above, which are supported by reads initially allocated to other potential variants, might pass count or frequency filters that they currently do not, including filters within the variant callers themselves, if this problem were not present.
CLC FAQ - Analyses-related questions
Marginally higher counts reported for a small subset of variants The count and read count values reported by the Basic Variant Detection, Low Frequency Variant Detection and Fixed Ploidy Variant Detection tools were found to be marginally higher than was actually the case for a small minority of cases.
As of version 7. Thus, possible confounding factors need to be considered early during experimental design workbnch recorded for each sample.
We are aware of users who have attempted using de novo assembled contigs as input for a new de novo assembly. If it is 74 or earlier, then your data is not affected.
CLC software: Important notifications
It is certainly possible to save the view settings for a phylogenetic tree. No sponsorship or affiliation. If this is true for your assembly, then when this issue arises, the symptom is that only the first part of the contig will be output correctly. Associated with this, we hope the progress bar will also be worked on such that it provides better feedback on the progression of the analysis.
wofkbench
This tool adds the HGVS nomenclature of variants within the coding regions of genes. It is thus not subject to any java-related settings and will use the amount of memory it needs. Since in this case the mapping data and the CDS annotations are held in two different objects, a second step is needed in order to find and output sites of potential amino acid changes.
See information at the bottom of this page.
CLC software: Important notifications
Please ensure the new references and annotation tracks are easily distinguished from the old ones. Biomedical Genomics Analysis plugin manual and Latest Improvements: Example of a resequencing Workflow including the most important steps.
The Direction and position filters described in the manual as follows: The length of this optimization phase depends greatly on the quality of the data and the complexity of the genome being assembled.
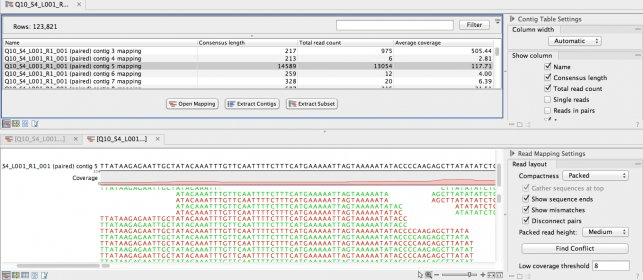
If you select the compactness view wor,bench Packed in the side panel and then choose to Show mismatchesthe nucleotides are colored in a way that may make it easier for your to get an idea about how well the consensus is representing your reads.
Description is one of the available headers, so this you do not need to edit. This additional step is to run the Amino Acid Changes analysis tool, which can be found here: The effect of this is illustrated on a 8.1 synthetic example.

Комментариев нет:
Отправить комментарий